Overview of variant annotation
Writers: @Do Minh Nguyet @Anh-Vu Mai-Nguyen
Advisor: @Thanh Nguyen
What is Variant Annotation?
Variant annotation is an important part of the analysis of genomic sequencing data. The annotation results can have a strong influence on the final conclusions of disease studies. Inaccurate or incomplete annotations can lead to missing potentially pathogenic DNA variants or diluting prominent variants in a series of false positives [1]. So what is variant annotation, and why is it important in the analysis of genomic data?
Variant annotation is the process of assigning functional information to DNA variants. Variant annotation provides us with information about variants, thereby supporting the analysis and interpretation of them. More specifically, we can examine the influence of aggregate associations of rare variants to identify, filter, and weight each component of that association (aggregation units). However, experiments involving the influence of rare variants still face some difficulties. The difficulties are largely caused by the scarcity of individuals carrying the variant allele. To optimize the calculation, rare variants in a biologically relevant region are often combined to form an aggregation unit. The types of variants and their relationships to the coding sequence in the genome provide us with information about the variation in the coding sequence and the variation in the gene product.
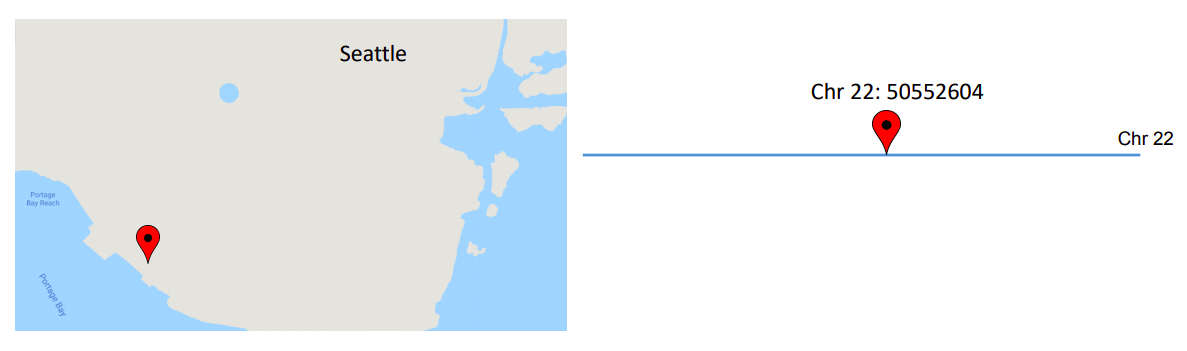
Annotating variants is like creating a road map for the genome [2]. To better understand variant annotation, let’s look at the following example:
Based on the map, the name of a city provides information about its location on the earth. Similarly, the chromosome and coordinates of the variant provide information about its location on the genome. Suppose we consider city A and the mutation on chromosome number 22 with coordinates 50552604.
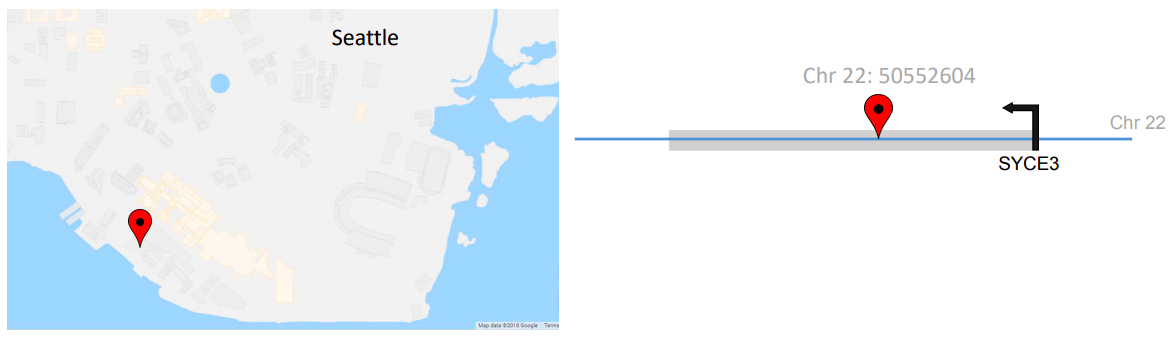
The gray outlines overlaid on the map indicate that you are in a building or structure. Similarly, the gene name annotation provides information about the gene that is overlaying the variation, such as the gene named SYCE3 in the image below.
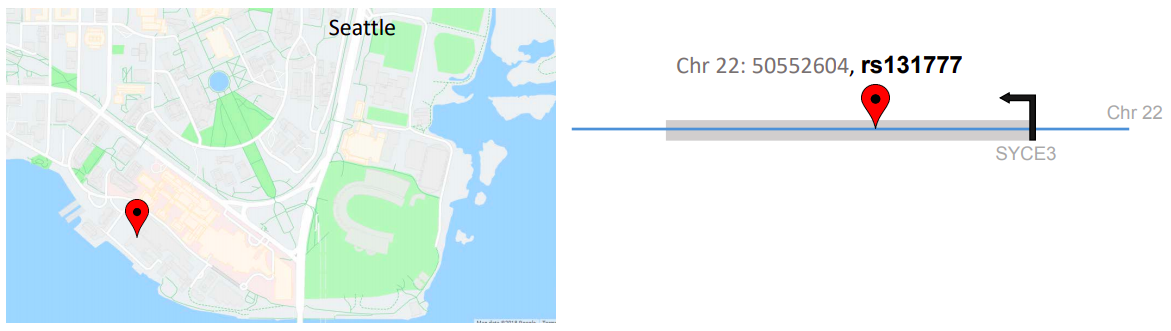
The roads indicate the possible paths to take from location A to any point B. Similarly, in the case under consideration, the identifier rs (Reference SNP cluster ID) and chú giải GWAS helped us determine that this variant had a prior relationship with a red blood cell trait called Mean corpuscular volume.
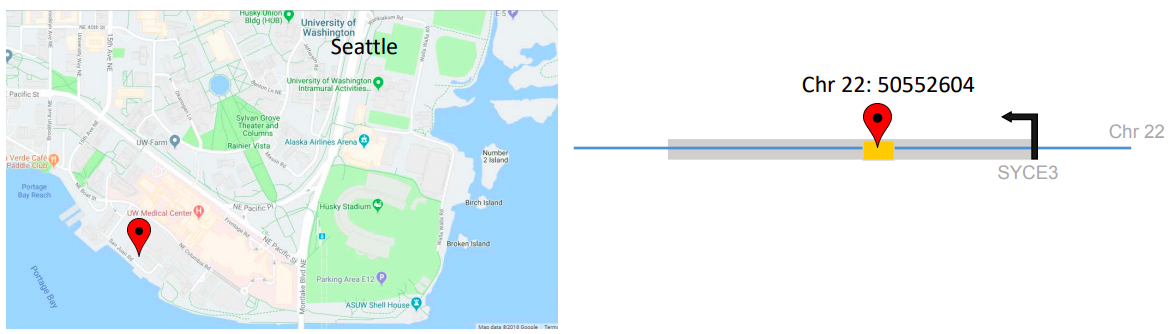
And finally, street and building names added to the map tell us exactly where we need to go. For example, we can walk to the UW farm, have lunch at Agua Verde, or go to Husky Stadium. Regulatory annotations help us identify variants that intersect according to certain rules, such as overlapping elements that are active in red blood cells and platelets, but not in brain or bladder cells.
Variant annotation support tools
The scientific community has developed many platforms and tools to support variant annotation. Some popular variant annotation support servers include: NCBI, Ensemble, UCSC, ENCylopedia Of DNA Elements (ENCODE), Roadmap Epigenomics Consortium, FANTOM5, dbSNP, etc.
Some popular open source variant annotation tools include: spliceAI, SIFT, PROVEAN, polyphen, MutationTaster, PHAST, Mutation Assessor, M-CAP, Linsight, GERP, GenoCanyon, FIRE, fathmm, CADD, VEP, etc.
SpliceAI
is an open-source artificial intelligence (AI) application software [4] announced by Illumina in 2019 [5]. SpliceAI annotates variants based on the connection of primary mRNA sequences. Using a deep learning network, spliceAI predicts junctions from a pre-mRNA transcript, setting the stage for accurate prediction of non-coding variants that cause aberrant splicing (cryptic splicing) [3]. Mutations located on non-coding segments are often overlooked in patients with rare genetic diseases, so the appearance of this software gives us more detailed information about those mutations in the gene.
SIFT
SIFT, SIFT was developed in 2011 and managed by Pauline Ng, is a tool that predicts whether amino acid substitutions affect protein function [8]. SIFT is widely used in bioinformatics, genetics, disease, and mutation studies. In 2017, a faster version of SIFT, SIFT 4G, was released, allowing users to scale up the computation and provide prediction data for more organisms. SIFT annotates and provides deleterious/tolerant predictions for single-point mutations. For insertion-deletion mutations (indels), SIFT only provides annotation results. Compared to SIFT, SIFT 4G – Sorting Intolerant From Tolerant For Genomes, is implemented on GPUs, so processing a protein takes only 3 seconds instead of 4 minutes [7].
PROVEAN
PROVEAN (Protein Variation Effect Analyzer) is a software released in 2012 with the purpose of predicting changes in biological function of proteins when there are point substitution or insertion-deletion mutations. PROVEAN works effectively in the case of filtering variant sequences to identify nonsynonymous or insertion-deletion variants that are predicted to be functionally important. The performance of PROVEAN can be comparable to popular tools such as SIFT or PolyPhen-2 [6].
Polyphen
Polyphen now known as PolyPhen-2 (Polymorphism Phenotyping v2) is a tool that predicts whether amino acid substitutions affect the structure and function of human proteins [9]. PolyPhen-2 was developed from PolyPhen for the purpose of annotating non-synonymous variants. Polyphen-2 relies on protein sequences, phylogenetic information, and structural information to annotate variants. The software considers whether the mutation is located in a region of the protein that is required for binding to other molecules to form secondary or tertiary structures. In particular, Polyphen-2 considers putative disulfide bonds, active sites, binding sites, and transmembrane domains and then performs calculations on a 3D model of the protein structure. Polyphen-2 also considers homologous proteins to see if the identified missense mutation is observed in other proteins of the same family [10].
MutationTaster
MutationTaster MutationTaster is a web-based variant annotation tool. MutationTaster evaluates variants in DNA sequences for their pathogenic potential. The software performs a series of in silico tests to estimate the effect of the variant on the gene product or protein. The tests are performed at the protein and DNA level, so MutationTaster is not limited to single amino acid substitutions but can also handle synonymous or intronic variants [11][12]. Mutation Taster is written in Perl and can process data from next generation sequencing (NGS) methods of all major platforms (Roche 454, Illumina Genome Analyzer and ABI SOLiD). MutationTaster uses a Naive Bayes classifier to decide whether the effect of all single variants is likely to be deleterious to the protein. The analysis results explain whether the change is a known or predicted pathogenic or harmless mutation and provide detailed information about the mutation [13]. The latest update of MutationTaster in 2021.
PHAST
Phylogenetic Analysis with Space/Time models – PHAST PHAST is best known as the search engine behind the conservation tracks in the University of California, Santa Cruz (UCSC) Genome Browser. PHAST is a freely available software package that includes command-line programs and supporting libraries for comparative and evolutionary genomics. PHAST also includes a number of tools for phylogenetic modeling, functional element identification, as well as utilities for manipulating genome alignments, trees, and annotations. The main subroutines of PHAST include phastCons (conservation scoring and identification of conserved elements), phyloFit (Determine the fit of phylogenetic models to aligned DNA sequences), phyloP (Calculate p-values for conservation or acceleration, lineage-specific or across all branches), phastOdds (Log-odds scoring for phylogenetic models or phylo-HMMs), exoniphy (Phylogenetic exon prediction), dless (Predict lineage-specific selected elements), prequel (Probabilistic reconstruction of ancestral sequences), and phastBias (Identify GC-biased gene conversion using phylo-HMMs) [14].
Mutation Assessor
Mutation Assessor predict the functional impact of amino acid substitutions in proteins, such as mutations found in cancer or missense polymorphisms. The functional impact is assessed based on the evolutionary conservation of the affected amino acid in protein homologs. The method has been validated on a large (60k) set of disease-associated polymorphisms (OMIM) and polymorphic variants [15]. The Mutation Assessor uses multiple sequence alignment (MSA), partitioned to reflect functional specificity, and generates a conservation score for each column to represent the functional impact of an aberrant variant. The Mutation Assessor generates individual MSAs using UniProt protein sequences. These are then partitioned based on UniProt and Pfam domain boundaries, and the 3D structure is used to generate matched family and subfamily sets [16]. Launched in 2011, Mutation Assessor has gone through four releases, the most recent in 2015.
M-CAP
Mendelian Clinically Applicable Pathogenicity (M-CAP), published in 2016 by the Bejerano lab at Stanford University, is the first pathogenicity classifier for rare missense variants in the human genome that has been tuned to the sensitivity required for clinical testing. M-CAP scores only rare missense variants: hg19, ENSEMBL 75 missense, ExAC v0.3 in which no metapopulation has a minor allele frequency above 1%. If a missense variant has no M-CAP score, the M-CAP prediction is assumed to be likely benign. M-CAP uses a gradient boosting tree classifier to learn a function of input features as a linear combination of decision trees, each of which is iteratively derived to correct previously misclassified elements [18].
Linsight
Linsight, developed in 2016, predicts non-coding nucleotide positions where mutations are likely to have severe physical consequences and are therefore likely to be phenotypically important. Linsight combines a generalized linear model for functional genomics data with a probabilistic model of molecular evolution. The method is fast and highly scalable, allowing it to exploit the “Big Data” available in modern genomics. In addition, Linsight was applied to the atlas of human enhancers and showed that health consequences at enhancers depend on cell type, tissue specificity, and constraints at the enhancers involved [19].
GERP
Genomic Evolutionary Rate Profiling – GERP released in 2011, identifies elements that are constrained in multiple alignments by quantifying substitution deficits. These deficits represent substitutions that would have occurred if the DNA element had been neutral, but did not occur because the element was functionally constrained. These deficits are called “rejected substitutions.” Rejected substitutions are a measure of natural constraint, reflecting the strength of past selection on the element [20].
GenoCanyon
GenoCanyon is an unsupervised statistical genome-wide functional annotation approach. GenoCanyon integrates genomic conservation measures and biochemical annotation data to predict the functional potential at each nucleotide. Using 22 computational and experimental annotations, the tool predicts the functional potential of each position in the human genome. With GenoCanyon, many of the same known functions can be predicted. Currently, with the latest update in 2015, the official GenoCanyon website [21] has available the prediction score for the entire human genome version hg19, the prediction score, and all 22 annotations [22].
FIRE
FIRE is a genome-wide variant annotation tool. FIRE assigns a higher score to SNVs that are more likely to alter the expression levels of neighboring genes. Because FIRE is specifically designed to regulate gene expression, the FIRE score does not directly correlate with pathogenicity or deleteriousness. Additionally, FIRE is specific to mRNA-level expression regulation and does not apply to SNVs that alter protein expression independently of mRNA expression [22]
fathmm
Functional Analysis through Hidden Markov Models – fathmm was developed in 2014 as a high-throughput web server capable of predicting the functional consequences of both coding variants, i.e. non-synonymous single nucleotide variants (nsSNVs), and non-coding variants in the human genome. For non-coding variant annotation, fathmm has two options including FATHMM-MKL (which uses the MKL algorithm that integrates functional annotations from ENCODE with nucleotide-based HMMs) and FATHMM-XF (which improves the accuracy in predicting the functional consequences of non-coding and coding single nucleotide variants (SNVs)) [23].
FATHMM-XF is a significant improvement over FATHMM-MKL. By using an expanded set of feature sets and an expanded set of models, the new method delivers higher accuracy than its predecessor on independent test sets. Like FATHMM-MKL, FATHMM-XF predicts whether single nucleotide variants in the human genome are likely to be functionally active in genetic diseases. FATHMM-XF uses separate models for coding and non-coding regions, to improve overall accuracy. Unlike FATHMM-MKL, FATHMM-XF models are built on a single-kernel dataset. The models learn the interactions between data sources to increase prediction accuracy across all regions of the genome [24][25]
CADD
CADD,
developed in 2014, is a tool to assess the deleteriousness of single nucleotide variants and insertion-deletions in the human genome. Although there are many variation scoring and annotation tools, most annotations tend to exploit a single type of information (e.g., conservation) and/or are limited in scope (e.g., false positives). CADD is a tool that integrates multiple annotations into one index by comparing variants that have survived natural selection with simulated mutations.
The C-score correlates strongly with allelic diversity, pathogenicity of both coding and non-coding variants, and experimentally measured regulatory effects, and also ranks causal variants highly in individual genome sequences. Finally, the C-scores of trait-associated intronic variants from genome-wide association studies (GWAS) were significantly higher than those from matched controls and correlated with study sample size, likely reflecting the increased precision of larger GWAS.
CADD can prioritize the quantification of functional, deleterious, and disease-causing variants across a wide range of functions, effect sizes, and genetic architectures and can be used to prioritize causal variants in both research and clinical settings [26].
VEP
VEP is a software suite for annotating and analyzing most forms of genetic variation in coding and non-coding regions of the genome. VEP is available as an online tool, command-line in Perl, and via the Ensembl REST API (transfer state representation) application program interface. Each interface is optimized to support different amounts of data and levels of bioinformatics experience. All three use the same core codebase to ensure consistent results across each interface. A comprehensive test suite supports all code, with continuous integration performed by Travis CI [27].
References:
[1] Choice of transcripts and software has a large effect on variant annotation – Genome Medicine
[2] https://si.biostat.washington.edu/sites/default/files/modules/variant_annotation_v1.pdf
[3] https://www.cell.com/cell/pdf/S0092-8674(18)31629-5.pdf
[4] spliceai
[5] Illumina Releases SpliceAI, Open Source AI software for Interpretation
[6] Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 7(10): e46688.
[7] https://sift.bii.a-star.edu.sg/sift4g/AboutSIFT4G.html
[8] Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC (2016) SIFT missense predictions for genomes. Nat Protocols 11: 1-9.
[9] Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. Nat Methods 7(4):248-249 (2010). PubMed PDF Supplemental Information
[10] https://bredagenetics.com/polyphenpolyphen2/
[11] Schwarz, Jana Marie; Rödelsperger, Christian; Schuelke, Markus; Seelow, Dominik (2010-08-01). “MutationTaster evaluates disease-causing potential of sequence alterations”. Nature Methods. 7 (8): 575–576. doi:10.1038/nmeth0810-575. ISSN 1548-7105. PMID 20676075.
[12] Schwarz, Jana Marie; Cooper, David N; Schuelke, Markus; Seelow, Dominik (2014-03-28). “MutationTaster2: mutation prediction for the deep-sequencing age”. Nature Methods. 11 (4): 361–362. doi:10.1038/nmeth.2890. ISSN 1548-7105. PMID 24681721
[13] Simcikova D, Heneberg P (December 2019). “Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases”. Scientific Reports. 9 (1): 18577. doi:10.1038/s41598-019-54976-4. PMC 6901466. PMID 31819097
[14] Ramani R, Krumholz K, Huang Y, Siepel A (2018) PhastWeb: a web interface for evolutionary conservation scoring of multiple sequence alignments using phastCons and phyloP, Bioinformatics, Volume 35, Issue 13, Pages 2320–232
[15] MutationAssessor.org /// functional impact of protein mutations
[16] Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: Applications to cancer genomics. Nucleic Acids Res. (2011)
[17] Jagadeesh, K., Wenger, A., Berger, M., Guturu, H., Stenson, P., Cooper, D., Bernstein, J., and Bejerano, G. (2016). M-CAP eliminates a majority of variants with uncertain significance in clinical exomes at high sensitivity. Nature Genetics, 2016. 48 (12) 1581 DOI: 10.1038/ng.3703
[18] Jagadeesh, K., Wenger, A., Berger, M., Guturu, H., Stenson, P., Cooper, D., Bernstein, J., and Bejerano, G. (2016). M-CAP eliminates a majority of variants with uncertain significance in clinical exomes at high sensitivity. Nature Genetics, 2016. 48 (12) 1581 DOI: 10.1038/ng.370
[19] Huang YF, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 2017;49(4):618-624. doi:10.1038/ng.3810
[20] Spies N, Weng Z, Bishara A, McDaniel J, Catoe D, Zook JM, Salit M, West RB, Batzoglou S, Sidow A. Genome-wide reconstruction of complex structural variants using read clouds. Nat Methods. 2017 Sep;14(9):915-920. doi: 10.1038/nmeth.4366. Epub 2017 Jul 17. PMID: 28714986; PMCID: PMC5578891.
[21] GenoCanyon Home
[22] Ioannidis NM, Davis JR, DeGorter MK, et al. FIRE: functional inference of genetic variants that regulate gene expression. Bioinformatics. 2017;33(24):3895-3901. doi:10.1093/bioinformatics/btx534
[23] Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, Day INM, Gaunt, TR. (2013). Predicting the Functional, Molecular and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum. Mutat., 34:57-65
[24] Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day INM, Gaunt TR, Campbell C (2014). An Integrative Approach to Predicting the Functional Consequences of Non-coding and Coding Sequence Variation. Bioinformatics 2015 May 15;31(10):1536-43.
[25] Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: enhanced accuracy in the prediction of pathogenic sequence variants via an extended feature set. (journal submission
[26] Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variant. Nat Genet. 2014 Feb 2. doi: 10.1038/ng.2892. PubMed PMID: 24487276.
[27] McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F.
The Ensembl Variant Effect Predictor. Genome Biology Jun 6;17(1):122. (2016). doi:10.1186/s13059-016-0974-4