Overview of the Polygenic Risk Score (PRS)
Advisor: @Dat Nguyen
Writer: @Do Minh Nguyet
Introduction
The Polygenic Risk Score (PRS) has been studied and circulated in the scientific community for many years. However, it was not until 2018 that it was demonstrated to have potential for use in clinical studies. A study by Amit V. Khera and colleagues at the Cardiovascular Disease Initiative of the Broad Institute in Cambridge, Massachusetts, identified people at high risk for five common diseases based on their genome alone [1].
The team used genome-wide association study (GWAS) data and an imputation method to assess millions of common genetic variants associated with coronary artery disease, atrial fibrillation, type 2 diabetes, inflammatory bowel disease, and breast cancer. For each disease, they applied a computational algorithm that combined information from all variants into a single number (PRS), reflecting a person’s genetic susceptibility to these diseases.
When the team tested their polygenic predictor of heart disease on 290,000 participants in the UK Biobank, they found that 8 percent of the population had a three-fold increased risk of heart attack. “We couldn’t have found that without genetic testing,” said Khera, who is also a cardiologist and geneticist at Massachusetts General Hospital. Using a similar approach for inflammatory bowel disease and breast cancer, the team found that their PRS consistently identified a group (between 2 and 10 percent of the population) at particularly high risk. Khera’s results provide compelling evidence in favor of more widespread use of PRS markers. The results from these studies show the benefits of early risk detection for disease prevention. A new question is how to develop the potential and improve the effectiveness of using polygenic markers in health settings. This article explores the development of genetic scores and approaches to addressing the challenges surrounding the use of these indices.
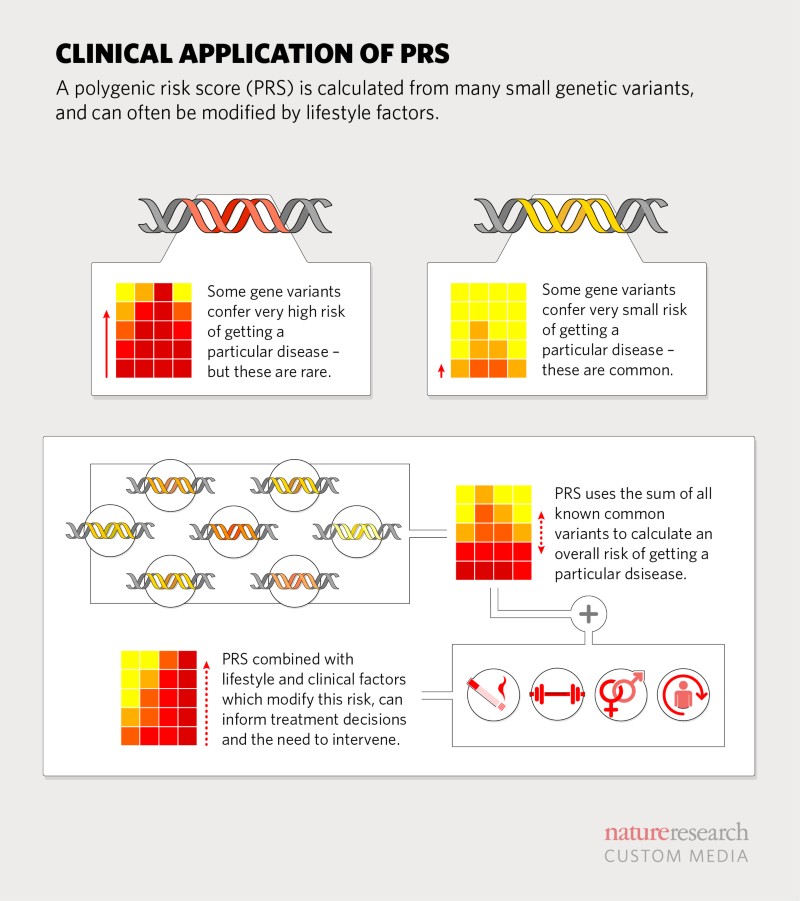
From single gene to genome-wide variations
Genetic testing is widely used to diagnose single-gene diseases, such as cystic fibrosis or sickle cell disease, which are caused by mutations in a single gene. These tests can also identify unaffected carriers of a disease-associated gene, allowing them to make more informed family planning decisions or, in cases such as Huntington’s disease, to plan in case they develop the condition in the future.
However, common diseases, such as type 2 diabetes and many neurodegenerative diseases, tend to be polygenic – influenced by a large number of genetic variants scattered throughout the genome, as well as environmental and lifestyle factors. New genomic technologies allow researchers to sequence genes quickly and inexpensively. Large gene panels, all of the protein-coding genes (exome) or the entire genome (whole genome sequencing, WGS), provide a complete survey of a person’s genetic makeup. For nearly two decades, geneticists have been comparing genomes, looking for differences that might explain why only some people develop certain diseases.
A genome-wide association study (GWAS) can identify such variation. These variations are often in the form of single nucleotide polymorphisms (SNPs). Through GWAS, researchers have found a large number of disease-associated variants, although the contribution of each variant is small. Over the years, they have developed tools to precisely determine the impact of a set of millions of disease-causing genetic variants [2].
Identifying a person’s susceptibility to specific diseases has many benefits. For example, Khera’s findings suggest that in women at high risk of breast cancer, starting screening earlier could improve outcomes. “Our goal is to empower people to overcome any predisposition to disease that is in their DNA,” says Khera. Even if some diseases are currently incurable, PRS could be used to match drugs in clinical trials. “A trial of an immunotherapy drug could be more profitable by focusing on people with high immune multifactor scores,” explains Desikan.
Improved polygenic index
An important concern surrounding the clinical implementation of PRS is that to date, scores have largely been calculated from European DNA sequences [3]. “The frequency and disease-relatedness of common genetic variants in African Americans is different from that in European Americans—and this reduces the accuracy of PRS,” says Martin. Martin is involved in several global projects aimed at characterizing genomic variation in diverse populations and developing statistical methods to analyze multiethnic data and improve the accuracy of PRS. One of these projects is the Neuropsychiatric Genetics in African Populations (NeuroGap). “The early human migrations out of Africa introduced genetic diversity into Europe, East Asia, and eventually the Americas,” she says. “Conducting large genetic studies in African populations would rapidly improve the accuracy of PRS for all populations.”
Because many health conditions are associated with environmental factors, lifestyle, and genetic susceptibility, combining PRS with other known risk factors would further improve risk prediction and aid in defining clinical thresholds [4]. According to Ali Torkamani, a geneticist at the Scripps Research Institute in La Jolla, California, there is sufficient evidence to support the use of PRS in decisions around statin therapy. For patients identified as having a clinically intermediate risk of developing coronary heart disease (from commonly measured clinical risk factors such as smoking, high blood pressure, and cholesterol levels), the addition of multifactorial risk information could help physicians make decisions about prescribing statins.
Using polygenic indices
Torkamani and his team developed MyGeneRank, an app that calculates an individual’s PRS for coronary heart disease from their 23andMe genetic data. Health data is collected on a mobile device using a series of questionnaires. Their goal is to understand how people respond to receiving their score and track any changes in health-related behaviors afterward.
Being informed about genetic risk can actually encourage the adoption of healthy lifestyle changes more broadly. The GeneRisk study in Finland presented at the 2018 ASHG conference showed that providing personalized cardiovascular risk information, based on a combination of traditional risk data and the PRS, helped promote healthy behaviors. Even participants at lower risk were inspired to lose weight, stop smoking, or see a doctor. Similar initiatives in Estonia, where the government is funding a genotyping programme for over 10% of the country’s population, are investigating the use of PRS for type 2 diabetes [5]. Individuals are given the option to find out their scores and those at highest risk are encouraged to make lifestyle changes, such as reducing sugar intake and increasing exercise, to prevent or delay the onset of diabetes.
Conclusion
Based on genetic data, PRS will soon be available to help people live healthier lives. “The great thing about DNA is that it is stable throughout your lifetime,” Khera says. “You can envision a not-too-distant future where, for $50, you get a genetic susceptibility report that identifies diseases you may be at risk for as a child, so you can take steps to prevent them.”
References
Article translated and edited from ![]() Polygenic risk: What’s the score? của Nature Portfolio.
Polygenic risk: What’s the score? của Nature Portfolio.
[1] Amit V. Khera et al. “Genome-wide polygenic scores for common diseasesidentify individuals with risk equivalent to monogenic mutations”. In:Na-ture Genetics(2018).doi:https://doi.org/10.1038/s41588- 018-0183-z.
[2] Julian R. Homburger et al. “Low coverage whole genome sequencing enablesaccurate assessment of common variants and calculation of genome-widepolygenic scores”. In:Genome Medicine(2019).doi:https://doi.org/10.1101/716977.
[3] Alicia R. Martin et al. “Clinical use of current polygenic risk scores mayexacerbate health disparities”. In:Nature Genetics(2019).doi:https://doi.org/10.1038/s41588-019-0379-x.
[4] Ali Torkamani, Nathan E. Wineinger, and Eric J. Topol. “The personaland clinical utility of polygenic risk scores”. In:Nature Reviews Genetics(2018).doi:https://doi.org/10.1038/s41576-018-0018-x.
[5] Kristi L ̈all MSc et al. “Personalized risk prediction for type 2 diabetes:the potential of genetic risk scores”. In:Genetics in Medicine(2017).doi:https://doi.org/10.1038/gim.2016.103.